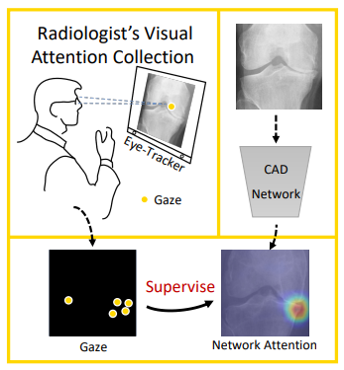
One of the most popular trends in medical image analysis is to apply deep learning techniques to computer-aided diagnosis (CAD). However, it is evident now that a large number of manually labeled data is often a must to train a properly functioning deep network. This demand for supervision data and labels is a major bottleneck, since collecting a large number of annotations from experienced experts can be time-consuming and expensive.
Continue reading